Morphological pre-processors¶
LBPM includes morphological pre-processing tools as utility functions. It is often useful to generate initial conditions for a 2-phase flow simulation based on a morphological approach. In particular, morphological tools can be used to provide a physical reasonable initial condition in cases where direct experimental observations are not available. These initial configurations are compatible with any of the 2-phase simulation protocols used by lbpm_color_simulator. These initialization approaches alter the fluid labels within the input files, writing a new file with the new morphologically assigned labels.
There are two main morphological pre-processors in LBPM
lbpm_morphdrain_pp– initialize fluid configuration based on morphological drainagelbpm_morphopen_pp– initialize fluid configuration based on morphological opening
Here we demonstrate lbpm_morphdrain_pp because it offers the most information. Although it is not perfect, the morphological drainage operation does a good job of approximating configurations observed along primary drainage processes performed under water-wet conditions. A limitation is that fluid trapped in the corners will not stop the morphological operation from eroding it. This should not discourage you too much – morphological tools are very practical and can save you a lot of time! It is also a good thing to be skeptical.
Since the morphological operation works on the input domain, associated parameters are added to the Domain section of the input file. Here we will set a target saturation Sw = 0.20, which will run the morphological drainage operation until the fluid labeled as 2 occupies 20% of the pore space or less. For the case considered in
example/DiscPack we specify the following information in the input file
Domain {
Filename = "discs_3x128x128.raw"
ReadType = "8bit" // data type
N = 3, 128, 128 // size of original image
nproc = 1, 2, 2 // process grid
n = 3, 64, 64 // sub-domain size
voxel_length = 1.0 // voxel length (in microns)
ReadValues = 0, 1, 2 // labels within the original image
WriteValues = 0, 2, 2 // associated labels to be used by LBPM
BC = 0 // fully periodic BC
Sw = 0.35 // target saturation for morphological tools
}
Once this has been set, we launch lbpm_morphdrain_pp in the same way as other parallel tools
mpirun -np 4 $LBPM_BIN/lbpm_morphdrain_pp input.db
Successful output looks like the following
Performing morphological opening with target saturation 0.350000
voxel length = 1.000000 micron
voxel length = 1.000000 micron
Input media: discs_3x128x128.raw
Relabeling 3 values
oldvalue=0, newvalue =0
oldvalue=1, newvalue =2
oldvalue=2, newvalue =2
Dimensions of segmented image: 3 x 128 x 128
Reading 8-bit input data
Read segmented data from discs_3x128x128.raw
Label=0, Count=11862
Label=1, Count=37290
Label=2, Count=0
Distributing subdomains across 4 processors
Process grid: 1 x 2 x 2
Subdomain size: 3 x 64 x 64
Size of transition region: 0
Media porosity = 0.758667
Initialized solid phase -- Converting to Signed Distance function
Volume fraction for morphological opening: 0.758667
Maximum pore size: 116.773801
1.000000 110.935111
1.000000 105.388355
1.000000 100.118937
1.000000 95.112990
1.000000 90.357341
1.000000 85.839474
1.000000 81.547500
1.000000 77.470125
1.000000 73.596619
1.000000 69.916788
1.000000 66.420949
1.000000 63.099901
1.000000 59.944906
1.000000 56.947661
1.000000 54.100278
1.000000 51.395264
1.000000 48.825501
1.000000 46.384226
1.000000 44.065014
1.000000 41.861764
1.000000 39.768675
1.000000 37.780242
1.000000 35.891230
1.000000 34.096668
1.000000 32.391835
0.575114 30.772243
0.433119 29.233631
0.291231 27.771949
Final void fraction =0.291231
Final critical radius=27.771949
Writing ID file
Writing file to: discs_3x128x128.raw.morphdrain.raw
The final configuration can be visualized in python by loading the output file
discs_3x128x128.raw.morphdrain.raw.
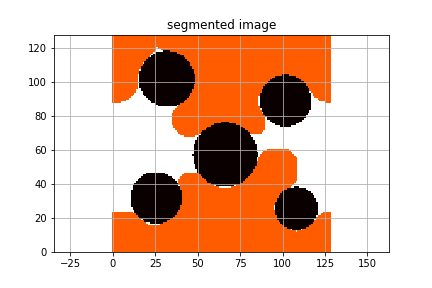
Fluid configuration resulting from orphological drainage algorithm applied to a 2D disc pack.¶